Artificial intelligence (AI) is an astonishingly rapidly growing technology and is already ‘changing the game’ in many fields. Together with Big Data, AI has been called the fourth industrial revolution. There is no single day when AI does not reveal itself in some form; from conversations about self-driving cars to the welcoming voice of Alexa on the Amazon smart speakers, Netflix making suggestions to watch 50 Shades of Grey and then Fitbit telling us to move more (I promise I will!). It is imperceptibly everywhere, and its applications will continue to expand to make our lives easier – benefitting us as long as we manage to keep AI within certain limits, we really don’t want to be like those poor ants described on the beneficial AI movement webpage!
A recent estimate from the Tufts Center for the Study of Drug Development shows that the cost to put a drug on the market is around $2.6 billion and more than 15 years of effort, which highlights the huge challenge the drug discovery industry is facing. Research and development (R&D) efficiency is decreasing and cost per drug is rising exponentially but sales are not. This is why R&D needs to come up with a strategy to accelerate innovation, and AI could be a good contender to convey at least part of what is needed. Augmenting the discovery process with advanced solutions seems to be indispensable at this point in time.
What is AI?
Let’s start by clarifying a few concepts about AI and Machine Learning (ML) which are not always clear in the mind of the less tech-savvy of us. AI is a subfield of computer science and can be defined as any algorithm or machine that, similarly to the human brain, shows intellectual abilities such as learning, evolving and in general understanding complex data. The expression was coined in 1956 by John McCarthy during his professorship at Dartmouth College in New Hampshire; in the same decade, the well-known Turing test designed to distinguish a human from a machine was conceived.
From a more philosophical point of view, we can separate AI into three broad types: a limited (or weak) AI, good at solving one specific problem, a general (or hard) AI with human-like intelligence (which does not exist yet) and a super AI with super human intelligence (the evil kind? It depends who you ask). The AI we are currently using is limited and it will be for some time.
Machine Learning (ML) is a subfield of AI and the term was introduced in the 1960s by International Business Machines (IBM), which refers to a numerical method that given certain data tries to find a way to explain the data and predict future data based on what it has learned.
Deep Neural Networks (DNNs) are a ML architecture lightly inspired by the human brain, comprised of a set of connected units (or neurons). DNNs are responsible for the rise of AI over the last few years. Differently from conventional ML, which usually reaches a learning plateau within a certain amount of data, Deep Learning (DL, a term created by combining DNN and ML) is data hungry: the more data you have, the better the performance – making it a very powerful and flexible ML tool. DNNs are not new, in fact they have been available in some form since the 1970s, but due to recent improvements in hardware performance at lower costs, availability of very large, curated data sets (Big Data), and key technical improvements (e.g. dropout), they have achieved unimaginable progress, especially in specific areas such as image and speech recognition.
How can AI help modernise drug discovery and drug design?
The applications of AI in the drug development process are already numerous, from target identification to selection of populations for clinical trials, drug repurposing and chemical syntheses planning. A key part of the discovery process is the design of the drug itself (this article refers to small-molecule drugs), which is fundamentally a multi-objective optimisation problem where several molecular properties need to be optimised simultaneously to meet specific goals such as potency, safety and metabolic stability. In the discovery phases, Lead Optimisation (LO) is the stage where most of this optimisation on a molecule generally happens and it is the most expensive phase in the identification of a drug candidate because hundreds of compounds are typically synthesised and tested in multiple assay systems. Solving this puzzle more rapidly and efficiently would mean enormous cost savings but also requires a deep understanding of very complex collections of data; a goal that can be intimidating. If there is one thing AI can do well, it is to make sense of very intricate data sets and therefore drug design seems the perfect domain of applicability.
Deep generative molecular design aims to create novel chemicals using deep learned AI technologies and, with the right architecture, perform multi-objective optimisation. In 2017 the Aspuru-Guzik group at Harvard published a seminal paper where they described for the first time a specific DNN architecture, a Variational Autoencoder (VAE), which is trained on numerous chemical structures and a few molecular properties of their choice which resulted in the ability to generate molecules which were chemically diverse with optimised properties from the input structures.
The VAE (Figure 1) described by Aspuru-Guzik is fundamentally a ’molecular optimizer’ composed of three different DNNs:
- an encoder that transforms a one-dimensional molecule representation (e.g. smiles) into a fixed-length vector stored in a latent space;
- a decoder that, starting from this latent representation, generates novel realistic synthetic samples and output their smiles code;
- a forecaster (multilayer perceptron – MLP) which predicts specific properties from the encoded molecule and through a Gaussian-based process, optimises them in the direction of preferred properties values.
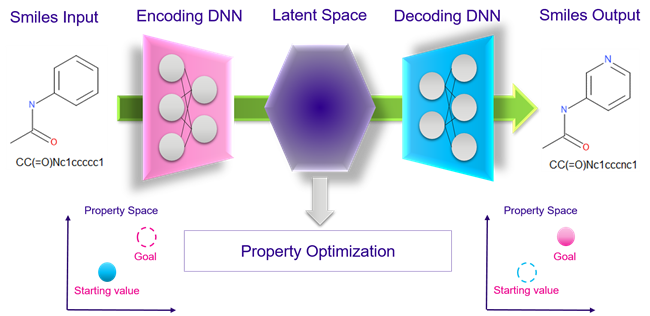
Figure 1. Neural network architecture of the variational autoencoder (VAE) proposed by Aspuru-Guzik.
Therefore, from an initial molecule, the trained generative VAE can output chemically diverse compounds with potentially improved properties and, in theory, with no limit on the number of properties that can be optimised.
Over the last two years since this influential work there have been at least 40 additional publications on the subject describing different and more complex DNN architectures capable of generating more realistic, synthesis-friendly and chemically diverse molecules which are better at optimising molecular properties. Another ML technique, deep reinforcement learning (DRL), seems particularly good at this last task due to its active learning based on constant feedback which does not need a lot of training data and is not biased by the training data. There is also a trend towards replacing one-dimensional molecular input (e.g. smiles) with three-dimensional chemical structures able to carry more information contained in the atoms’ spatial arrangement with a consequent increase in prediction accuracy.
We think that simplifying the access to these technologies and improving their interpretability is the key to increasing their use, utility and eventually, acceptance. In addition, their implementation should be pursued through modern data analysis environments (e.g. Knime and Pipeline Pilot) familiar to the drug designer, which will simplify the design of advanced data analysis workflows and eliminate the requirement of particular coding skills. In this way, more users would be able to effortlessly create, use and modify multifaceted analytics protocols to fit their needs. Therefore, users could focus mainly on the only area of competitive advantage: the data. Data create real value, algorithms don’t, and to use them fully, AI competences need to be grasped in-house.
Based on the initial results and constant developments in AI, a new frontier for intelligent molecular generation and optimisation could emerge, placing groups that can master it at the forefront for novel intellectual property generation and potentially new discoveries. At this point in time it is difficult to say if deep generative models will be a game-changer in drug design, but we believe they are here to stay and will be another tool in the hands of the drug designer.
Comments
If you are a British Pharmacological Society member, please
sign in to post comments.